By Dan Rubinstein
Antibiotics have been a staple in the animal production industry since the 1950s. They’re used to treat infections, prevent illnesses and enhance growth in cattle, pigs and poultry, helping companies bring large quantities of meat to market at affordable prices.
With increasing awareness and public concern about pathogens evolving resistance to antibiotics, livestock producers began moving away from “medically important antibiotics” in the 1990s, switching to medications that aren’t suitable for humans.
Problem solved? Not so fast.
Carleton University evolutionary biology researcher Alex Wong published a paper in 2019 suggesting that the use of antibiotics not considered important to human health on animals could still lead to antimicrobial resistance.

Carleton University evolutionary biology researcher Alex Wong
That paper caught the attention of Perdue Farms, one of the biggest poultry producers in the United States. The company, which also has pork, beef and lamb brands and does not use antibiotics in its operations, contacted Wong and asked if they could help support a follow-up study.
Published recently in the American Society for Microbiology’s mSphere Journal, Wong’s new research, co-authored by PhD candidate Asalia Ibrahim and recent graduate Jason Au, affirms that ionophores — a class of antibiotics widely used in animal production — could have long-term implications for human health.
“Despite the fact that we don’t use ionophores in humans, there are still risks,” says Wong.
“Our study indicates that we should hold back on the assumption that we can use ionophores whenever we want. There’s potential for resistance to antibiotics used in humans and we need to be more attentive.
“This research underscores the importance of understanding the interconnectedness of animal and human health.”
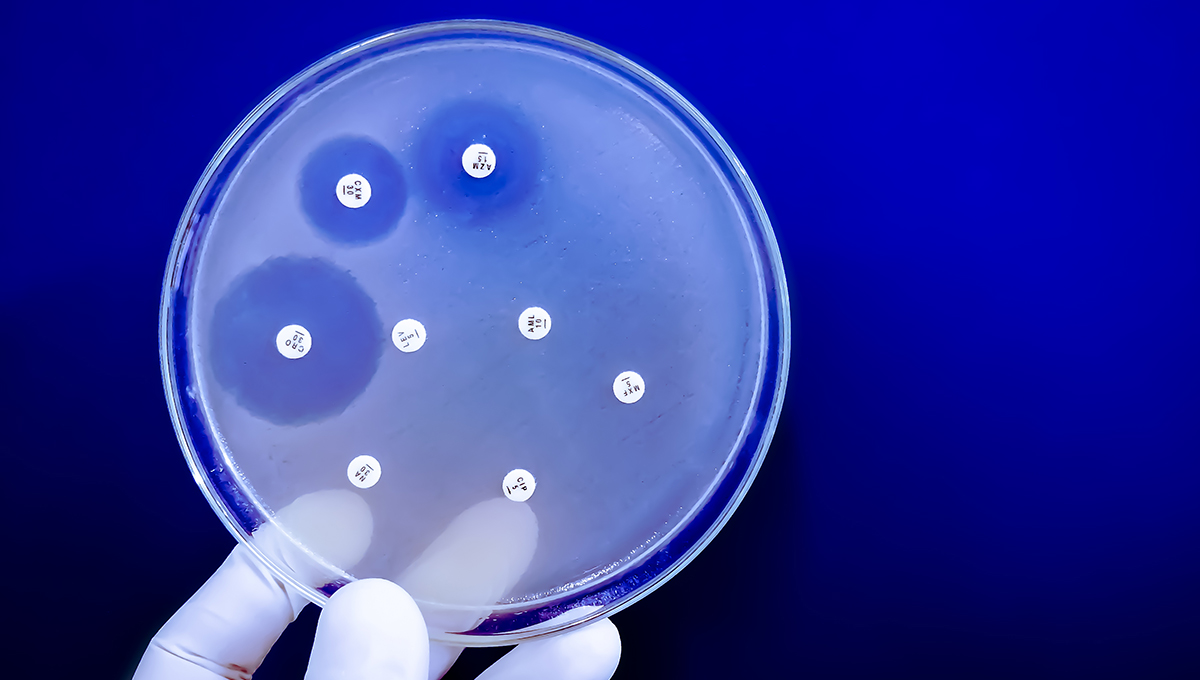
How Bacteria Adapt to New Environments
Working out of the Experimental Molecular Evolution Lab at Carleton, Wong’s research team primarily explores bacterial evolution.
He’s interested in how bacteria adapt to new environments, such as new food sources, and in more applied questions like how bacteria evolve resistance to antibiotics.
People can acquire antibiotic resistant infections from eating meat that’s been treated with meds, through farmers who’ve been in contact with animals, or from environmental contamination in water and other substances.
Wong’s 2019 paper noted that there can be associations between the resistance gene for ionophores and the resistance gene for vancomycin, which is prescribed by doctors to treat severe bacterial infections, in particular hospital-acquired infections.
In the half dozen years between then and now, among the vast amounts of data uploaded to public databases, researchers have shared genetic sequences from myriad pathogens isolated from people, from animals and from the environment.
None of this data was collected with ionophores in mind, Wong explains, but he can zoom in on the sequences that contain ionophore resistance genes and determine how many are linked to genes for resistance to medically important antibiotics.
“We found over 2,000 cases where bacterial isolates have resistance to these particular ionophores,” he says.
“We found them all over the world, on every continent except Antarctica, and it’s quite common for them to be linked to genes for medically important resistance. We didn’t even have to generate any new sequence data. We just had to sift through and analyze what was already available.”
Responsible Antibiotic Use
Perdue Farms is using this new information to inform discussions about responsible antibiotic use in animal agriculture.
Although he doesn’t want to make predictions, Wong points out that while most big poultry manufacturers have stopped using antibiotics, many are starting to use ionophores again.
“I would hope that this causes some of these companies to think twice,” he says.
“I think there’s the potential for antibiotic resistance to become a bigger problem. Many factors contribute to the spread of resistance in humans and it looks like this is becoming one of them.”
Beyond this project, Wong recently received funding from the Canadian Institutes of Health Research for more work on antibiotic resistance.
During the COVID-19 pandemic, his team was part of a large collaborative effort that developed a protocol for swabbing the floors of hospitals and long-term care homes to predict outbreaks by determining how much of the virus was present — much like picking up on the coronavirus signal from wastewater sampling.
Now they’ll be swabbing hospital floors to look for antibiotic resistance genes.
“We’ll be asking if what we find mirrors the patient load of antibiotic resistance in the hospital,” says Wong.
“We want to find out if this can be an effective surveillance tool for antibiotic resistance and to deepen our understanding of patient communities.”
During the pandemic, Wong also did research on using ultraviolet light to decontaminate masks and other personal protective equipment.
This may seem like more diverse areas of inquiry than one sees in a typical lab, but they’re all related to genetics, just applied to different organisms.
“There’s a long tradition of evolutionary biologists not being wedded to a particular system,” Wong says. “The really foundational questions are driven by curiosity and wonder, and our lab wants to apply some of these concepts in a concrete way. It’s a nice balance and I’m certainly never bored.”
Thursday, June 26, 2025 in Biology, Faculty of Science, Health
Share: Twitter, Facebook



